Postdoctoral Research Fellow
Pholpat (Big) Durongbhan
I develop reproducible quantitative methods for biomedical imaging and time-series data. My work builds high-resolution CT and synchrotron pipelines for osteochondral morphometrics, alongside signal-analysis frameworks spanning EEG and acoustics. I focus on validated feature extraction, interpretable modelling of structure–function relationships, and workflow automation that turns complex datasets into clinically meaningful metrics.

What are my research interests?
Research Highlights
As an interdisciplinary researcher at the intersection of engineering, biomedicine, and computer science, I specialise in developing quantitative image analysis tools for reproducible and high-precision biomedical assessments. In my BEng (Thailand), MSc (UK), PhD (Australia), and postdoctoral roles (Australia, Japan). I work at the intersection of engineering, biomedicine, and computer science to build quantitative analysis tools for high-precision, reproducible biomedical assessment.
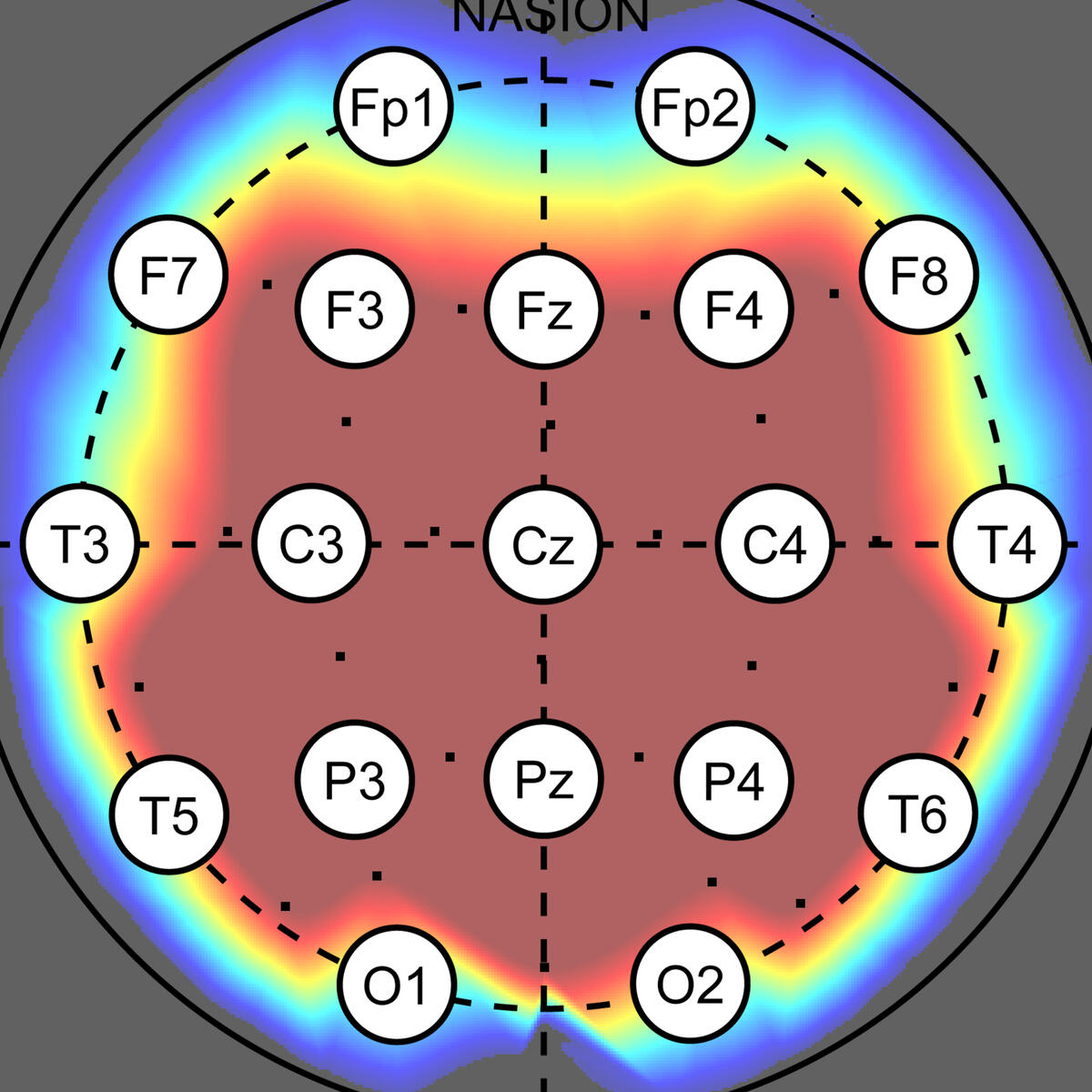
Assessment of Brain Connectivity
○ Developed machine-learning and non-linear signal analysis tools for dementia diagnosis and brain connectivity profiling (Cranfield University, Royal Hallamshire Hospital).○ Built reproducible feature-extraction and classification workflows to support robust interpretation of EEG connectivity markers.○ Produced two high-impact publications and continuing work on how sampling frequency influences machine learning classification accuracy.
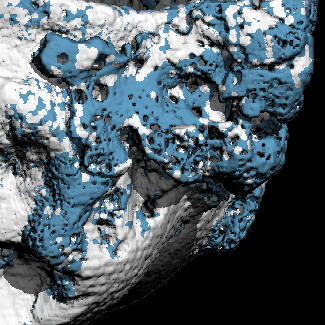
Morphological Analysis of Joint Tissues
○ Developed analysis workflows to quantify structural changes in joint tissues using contrast-enhanced and synchrotron-radiation microCT (University of Melbourne).○ Designed pipelines for reproducible morphometrics across different species and experimental protocols, with careful validation of measurement stability.○ Demonstrated robustness across contrast agents and imaging resolutions, enabling adaptable deployment in diverse osteoarthritis and joint-tissue studies.
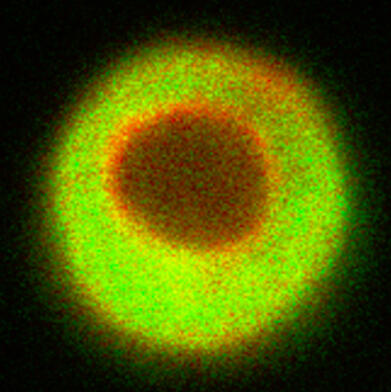
Quantification of Cell and Matrix Morphology
○ Built image-processing pipelines for quantitative analysis of cell and matrix in cartilage tissue constructs from 3D microscopy data (Erasmus MC, Murdoch Children’s Research Institute, University of Melbourne).○ Addressed common microscopy artifacts, including depth-wise intensity attenuation and ambiguous boundaries, to improve segmentation reliability.○ Enabled precise volumetric quantification of matrix deposition for comparative analysis across conditions and experimental batches.
What funding have I secured?
Selected Research Projects & Fellowships
In addition those listed, I have received multiple competitive postdoctoral fellowship offers and collaborative facility-access awards.
2026 JSPS Postdoctoral Fellowship - Standard Program
Establishing image-driven estimation of embryonic mechanical properties for reproductive health.
2024 University of Melbourne Early Career Researcher Grant
High-throughput, high-resolution image analysis platform for quantitative arthritis research.
2023 ANZBMS Bone Health Foundation Grant
Remodelling of the osteochondral interface using contrast-enhanced micro-computed tomography
2023 Australian Synchrotron Beamtime Grant
Nanoscale Measurement of Porous Channels in the Bone-cartilage Interface of a Mouse Model of Osteoarthritis using Synchrotron Radiation Phase-Contrast Imaging
What have I published?
Scholarly Works
10. Q1 P. Durongbhan, C. E. Davey, K. S. Stok, “Empirical Modelling Workflow for Resolution Invariant Assessment of Osteophytes”, IEEE Transactions on Biomedical Engineering, December 2024. DOI: 10.1109/TBME.2024.34316349. Software paper M. T. Kuczynski, N. J. Neeteson, K. S. Stok, A. J. Burghardt, M. A. Espinosa Hernandez, J. Vicory, J. J. Tse, P. Durongbhan, S. Bonaretti, A. K. O. Wong, S. K. Boyd, S. L. Manske, “ORMIR_XCT: A Python package for high resolution peripheral quantitative computed tomography image processing”, The Journal of Open Source Software, 9(97), 6084, May 2024. DOI: 10.21105/joss.060848. Book chapter; quantitative analysis of Thai prosody P. Tiratanti, & P. Durongbhan, “Reported thought embedded in reported speech in Thai news reports.” in The Grammar of Thinking: From Reported Speech to Reported Thought in the Languages of the World (pp. 239–260). De Gruyter Mouton, 2023. DOI: 10.1515/9783111065830-0097. Q1 H. Liu, P. Durongbhan, C. E. Davey, K. S. Stok, “Image Registration in Longitudinal Bone Assessment using Computed Tomography”, Current Osteoporosis Reports, June 2023. DOI: 10.1007/s11914-023-00795-66. Field journal; launched 2021 P. Durongbhan, J. W. MacKay, J. E. Schadow, C. E. Davey, K. S. Stok. “Quantitative morphometric analysis in tibiofemoral joint osteoarthritis imaging: a literature review”, Osteoarthritis Imaging, vol. 3, issue 1, 100088, March 2023. DOI: 10.1016/j.ostima.2023.1000885. Q1 P. Durongbhan, M. O. Silva, Z. Li, N. Ansari, R. Y. N. Kour, C. E. Davey, K. S. Stok, “A microCT imaging protocol for reproducible and efficient quantitative morphometric analysis (QMA) of joint structures for in situ mouse tibio-femoral joint”, Bone, vol. 166 (116606), January 2023. DOI: 10.1016/j.bone.2022.1166064. Q1 P. Durongbhan, C. E. Davey, K. S. Stok, “SPHARM-PDM Based Image Preprocessing Pipeline for Quantitative Morphometric Analysis (QMA) for in situ Joint Assessment in Rabbit and Rat Models,” Scientific Reports, 12 (1), January 2022. DOI: 10.1038/s41598-021-04542-83. Q1 B. A. Besler, J. E. Schadow, P. Durongbhan, T. H. Steiner, R. J. Choo, M. A. Zulliger, M. Wilke, K. Atal, C. Firminger, A. Quintin, B. Koller, R. Müller, D. Nesic, K. S. Stok, “Quantitative Measures of Bone Shape, Cartilage Morphometry and Joint Alignment are Associated with Disease in an ACLT and MMx Rat Model of Osteoarthritis,” Bone, vol. 146 (115903), May 2021. DOI: 10.1016/j.bone.2021.1159032. Q1; top 15% most cited on Scopus Y. Zhao, Y. Zhao, P. Durongbhan, L. Chen, S. A. Billings, P. Zis, Z. C. Unwin, M. D. Marco, A. Venneri, D. J. Blackburn, and P. G. Sarrigiannis, “Imaging of Nonlinear and Dynamic Functional Brain Connectivity Based on EEG Recordings with the Application on Diagnosis of Alzheimer's Disease,” IEEE Transactions on Medical Imaging, vol. 39, issue 5, pp. 1571-1581, May 2020. DOI: 10.1109/TMI.2019.29535841. Q1; top 5% most cited on Scopus P. Durongbhan, Y. Zhao, L. Chen, P. Zis, M. De Marco, Z. C. Unwin, A. Venneri, X. He, S. Li, Y. Zhao, D. J. Blackburn, and P. G. Sarrigiannis, “A Dementia Classification Framework Using Frequency and Time-Frequency Features Based on EEG Signals,” IEEE Transactions on Neural Systems and Rehabilitation Engineering, vol. 27, no. 5, pp. 826–835, May 2019. DOI: 10.1109/TNSRE.2019.2909100
What have my journey been like?
My Journey Thus Far
I received my initial training in Electrical Engineering, with a focus on signal processing and systems analysis, at Chulalongkorn University (Thailand). This foundation shaped how I approach complex data problems—emphasising mathematical rigor, reproducibility, and interpretable modelling.I later completed a Master’s degree at Cranfield University (UK), where I worked on high-resolution EEG-based machine-learning methods for dementia diagnosis, in close collaboration with clinical neurologists. This experience introduced me to translational research at the interface of engineering, data science, and medicine.During my PhD and postdoctoral work at the University of Melbourne (Australia), my research shifted toward high-resolution biomedical imaging, where I developed reproducible analysis pipelines for bone–cartilage and joint tissues using microCT and synchrotron imaging. Across these stages, my work has been united by a common goal: building robust quantitative methods that translate complex imaging and time-series data into meaningful biological and clinical insight.
© Pholpat Durongbhan. All rights reserved.